|
||||||||||||
|
About mentha
mentha archives evidence collected from different sources and presents these data in a complete and comprehensive way. Its data comes from manually curated protein-protein interaction databases that have adhered to the IMEx consortium. The aggregated data forms an interactome which includes many organisms. mentha is a resource that offers a series of tools to analyse selected proteins in the context of a network of interactions. Protein interaction databases archive protein-protein interaction (PPI) information from published articles. However, no database alone has sufficient literature coverage to offer a complete resource to investigate "the interactome". mentha's approach generates every week a consistent interactome (graph). Most importantly, the procedure assigns to each interaction a reliability score that takes into account all the supporting evidence. mentha offers eight interactomes (Homo sapiens, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Escherichia coli K12, Mus musculus, Rattus norvegicus, Saccharomyces cerevisiae) plus a global netowrk that comprises every organism, including those not mentioned. The website and the graphical application are designed to make the data stored in mentha accessible and analysable to all users. Go to User guide Our service displays content extracted from different sources. This content is responsibility of the entity that makes it available.
Reference
Please, in any article making use of the data extracted from mentha, refer to: mentha: a resource for browsing integrated protein-interaction networks Alberto Calderone, Luisa Castagnoli & Gianni Cesareni Nature Methods 10, 690 (2013). doi:10.1038/nmeth.2561
Source databases 




mentha integrates these databases because of their characteristics. They all use the same curation policies and the same complex expansion model. Acknowledgment
Statistics for main model organisms other organisms are available Last update: 15th February 2026
User guideHow to search Search overview
Search example Search all elongation factors in arabidopsis thaliana
ALTERNATIVE
Result page
List page 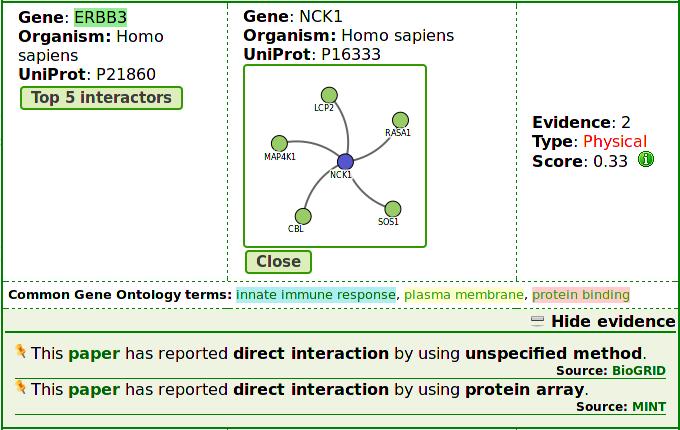
Cytoscape and mentha 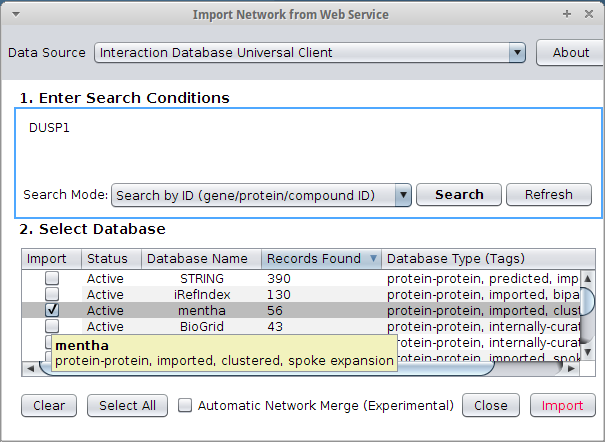 Once you have queired you proteins Cytoscape will assemble a network for you with all the details you need: experimental methods, references to articles and so on. 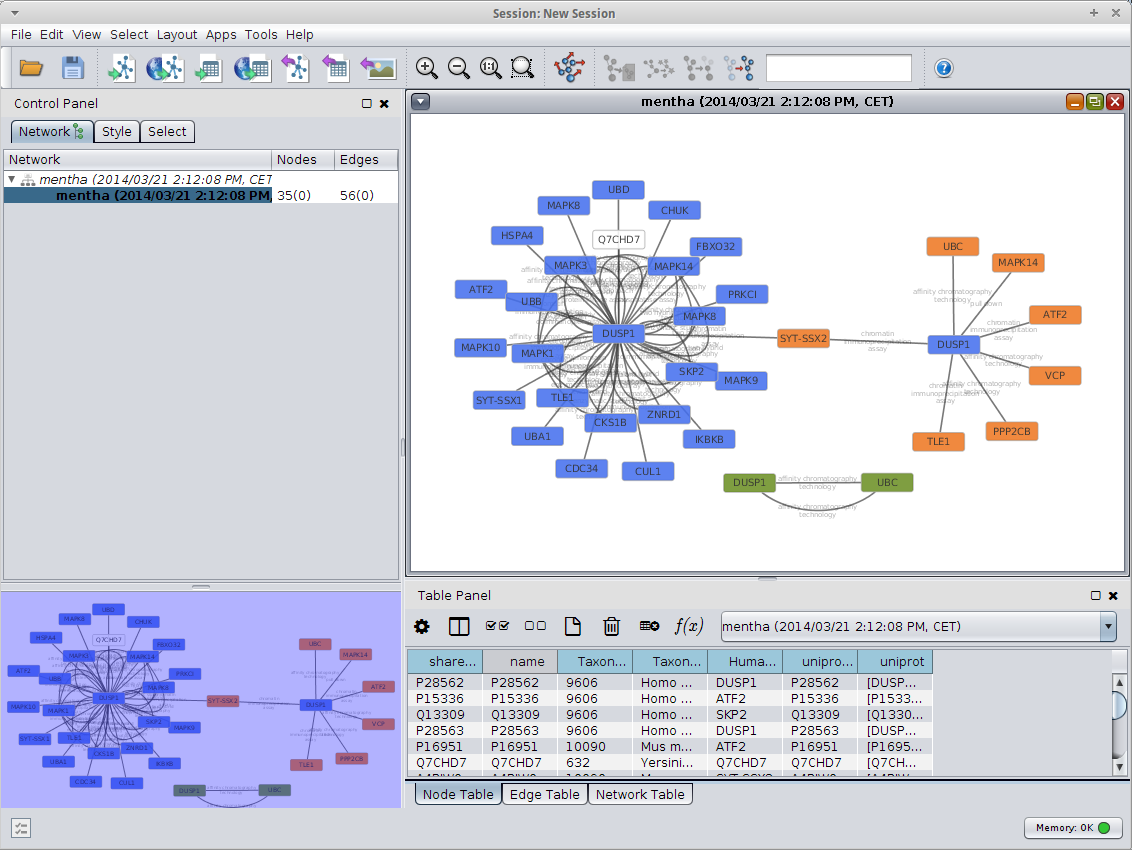 |
||||||||||||
| University of Tor Vergata, Rome - Italy |



